Transcriptomic analysis of host immune response in the chickens infected by avian leukosis virus J using RNA-Seq
Keywords:
Differential gene expression analysis, Immune system, Gallus Gallus, Avian leukosis viruses, RNA-seqAbstract
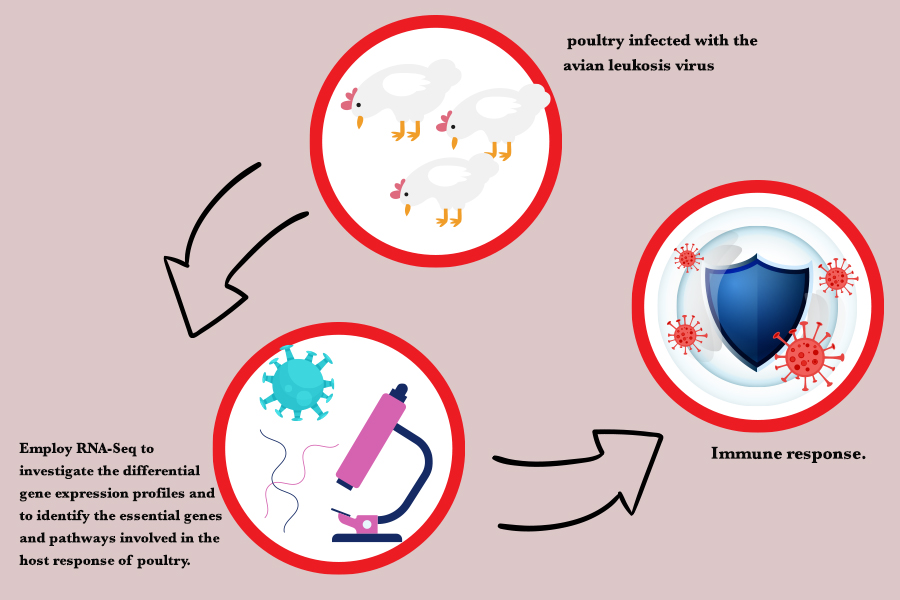
The poultry's immune system is regulated by various genes involved in both innate and acquired immune responses. Understanding the changes in gene expression post-infection is essential for developing effective intervention strategies and improving disease management in the poultry industry. This study aimed to employ RNA-Seq to investigate the differential gene expression profiles and to identify the essential genes and pathways involved in the host response of poultry infected with the avian leukosis virus. For this purpose, RNA-Seq data of healthy and avian leukosis virus-infected birds on days 24 (n=6) and 40 (n=6) post-infection were used. After quality control and preprocessing, we aligned the reads to the chicken reference genome using STAR software and quantified gene expression using HTSeq-Count. Differential gene expression analysis was performed using the edgeR package in R. The results of this study showed that the uniquely mapped read percentage ranged from 78.07% to 87.74%, and the mismatch rate per base was found to vary between 0.77% and 1.45%. A total of 2,213 and 1165 genes exhibited significant differential expression compared to the control group on day 24 and 40 post-infection, respectively (p<0.05). The gene ontology enrichment and pathway analysis revealed that six candidate genes, AvBD1, AvBD6, CATH1, CATH2, CATH3, and DEFB4A, are associated with the immune response on days 24 and 40. Additionally, on day 24, two more candidate genes, AvBD5 and LYG2, were found to be involved in the immune response.
Downloads
Downloads
Published
Issue
Section
License
Copyright (c) 2024 Paria Azamian, Saheb Foroutanifar, Alireza Abdolmohammadi (Author)

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.